Introduction to Pumas
This is an introduction to Pumas, a software system for pharmaceutical modeling and simulation. Pumas supports NLME (non-linear mixed effects models) estimation and simulation, NCA (non-compartmental analysis), Bioequivalence analysis, optimal design, and more.
In this tutorial we focus on NLME. The basic workflow of NLME with Pumas is:
- Build a model.
- Define subjects or populations to simulate or estimate.
- Analyze the results with post-processing and plots.
We will show how to build a multiple-response PK/PD model via the @model macro, define a subject with single doses, and analyze the results of the simulation. Fitting of data using any of the methods available in Pumas is showcased in the tutorials. This tutorial is designed as a broad overview of the workflow and a more in-depth treatment of each section can be found in the tutorials.
Working Example
Let us start by showing complete simulation code, and then break down how it works.
using Pumas
using PumasUtilities
using CairoMakie######### Turnover model
inf_2cmt_lin_turnover = @model begin
@param begin
tvcl ∈ RealDomain(; lower = 0)
tvvc ∈ RealDomain(; lower = 0)
tvq ∈ RealDomain(; lower = 0)
tvvp ∈ RealDomain(; lower = 0)
Ω_pk ∈ PDiagDomain(4)
σ_prop_pk ∈ RealDomain(; lower = 0)
σ_add_pk ∈ RealDomain(; lower = 0)
# PD parameters
tvturn ∈ RealDomain(; lower = 0)
tvebase ∈ RealDomain(; lower = 0)
tvec50 ∈ RealDomain(; lower = 0)
Ω_pd ∈ PDiagDomain(1)
σ_add_pd ∈ RealDomain(; lower = 0)
end
@random begin
ηpk ~ MvNormal(Ω_pk)
ηpd ~ MvNormal(Ω_pd)
end
@pre begin
CL = tvcl * exp(ηpk[1])
Vc = tvvc * exp(ηpk[2])
Q = tvq * exp(ηpk[3])
Vp = tvvp * exp(ηpk[4])
ebase = tvebase * exp(ηpd[1])
ec50 = tvec50
emax = 1
turn = tvturn
kout = 1 / turn
kin0 = ebase * kout
end
@init begin
Resp = ebase
end
@vars begin
conc := Central / Vc
edrug := emax * conc / (ec50 + conc)
kin := kin0 * (1 - edrug)
end
@dynamics begin
Central' = -(CL / Vc) * Central + (Q / Vp) * Peripheral - (Q / Vc) * Central
Peripheral' = (Q / Vc) * Central - (Q / Vp) * Peripheral
Resp' = kin - kout * Resp
end
@derived begin
dv ~ @. Normal(conc, sqrt((conc * σ_prop_pk)^2 + σ_add_pk^2))
resp ~ @. Normal(Resp, σ_add_pd)
end
end
turnover_params = (;
tvcl = 1.5,
tvvc = 25.0,
tvq = 5.0,
tvvp = 150.0,
tvturn = 10,
tvebase = 10,
tvec50 = 0.3,
Ω_pk = Diagonal([0.05, 0.05, 0.05, 0.05]),
Ω_pd = Diagonal([0.05]),
σ_prop_pk = 0.15,
σ_add_pk = 0.5,
σ_add_pd = 0.2,
);regimen = DosageRegimen(150; rate = 10, cmt = :Central)
pop = map(i -> Subject(; id = i, events = regimen), 1:10)Population
Subjects: 10
Observations: sd_obstimes = [
# single dose observation times
0.25,
0.5,
0.75,
1,
2,
4,
8,
12,
16,
20,
24,
36,
48,
60,
71.9,
];sims = simobs(inf_2cmt_lin_turnover, pop, turnover_params; obstimes = sd_obstimes)Simulated population (Vector{<:Subject})
Simulated subjects: 10
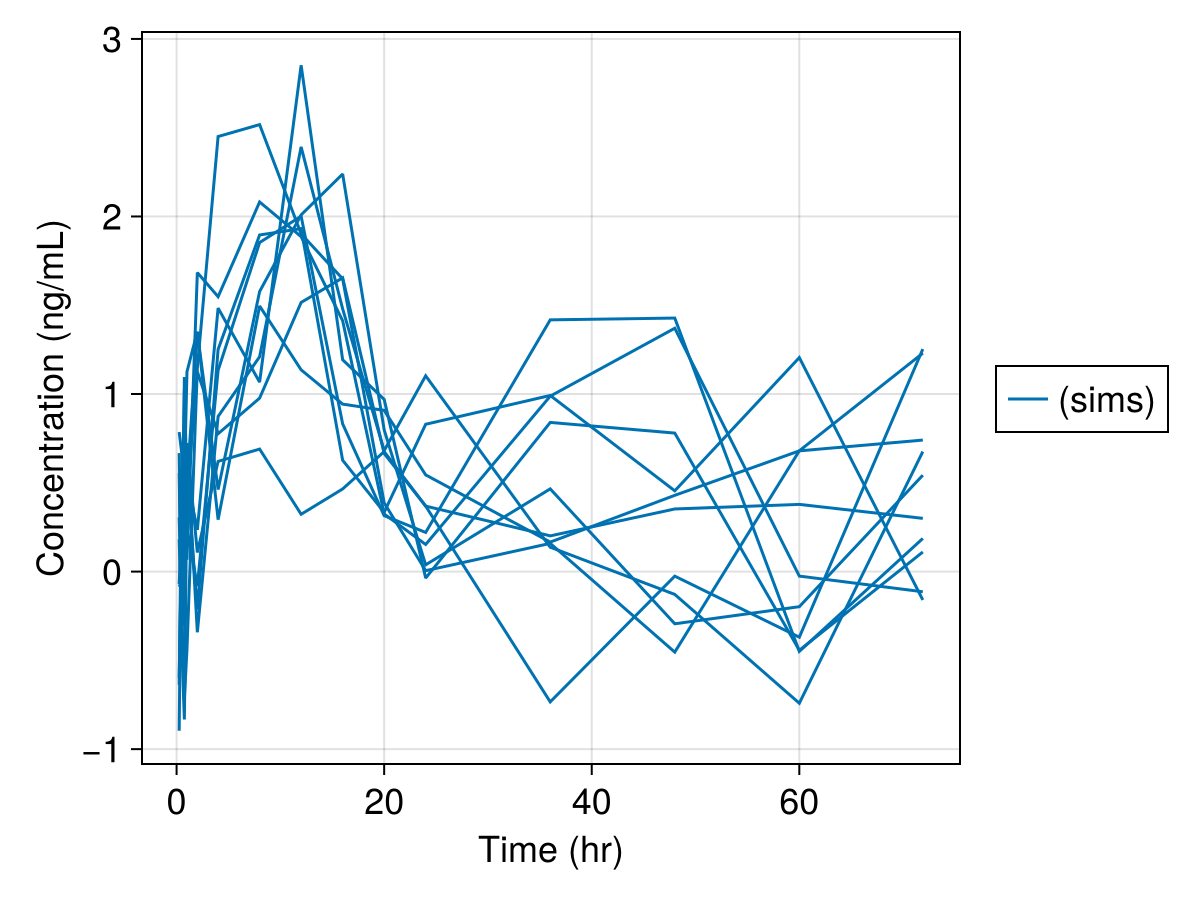
Simulated variables: dv, respsim_plot(
inf_2cmt_lin_turnover,
sims;
observations = [:dv],
figure = (; fontsize = 18),
axis = (; xlabel = "Time (hr)", ylabel = "Concentration (ng/mL)"),
)
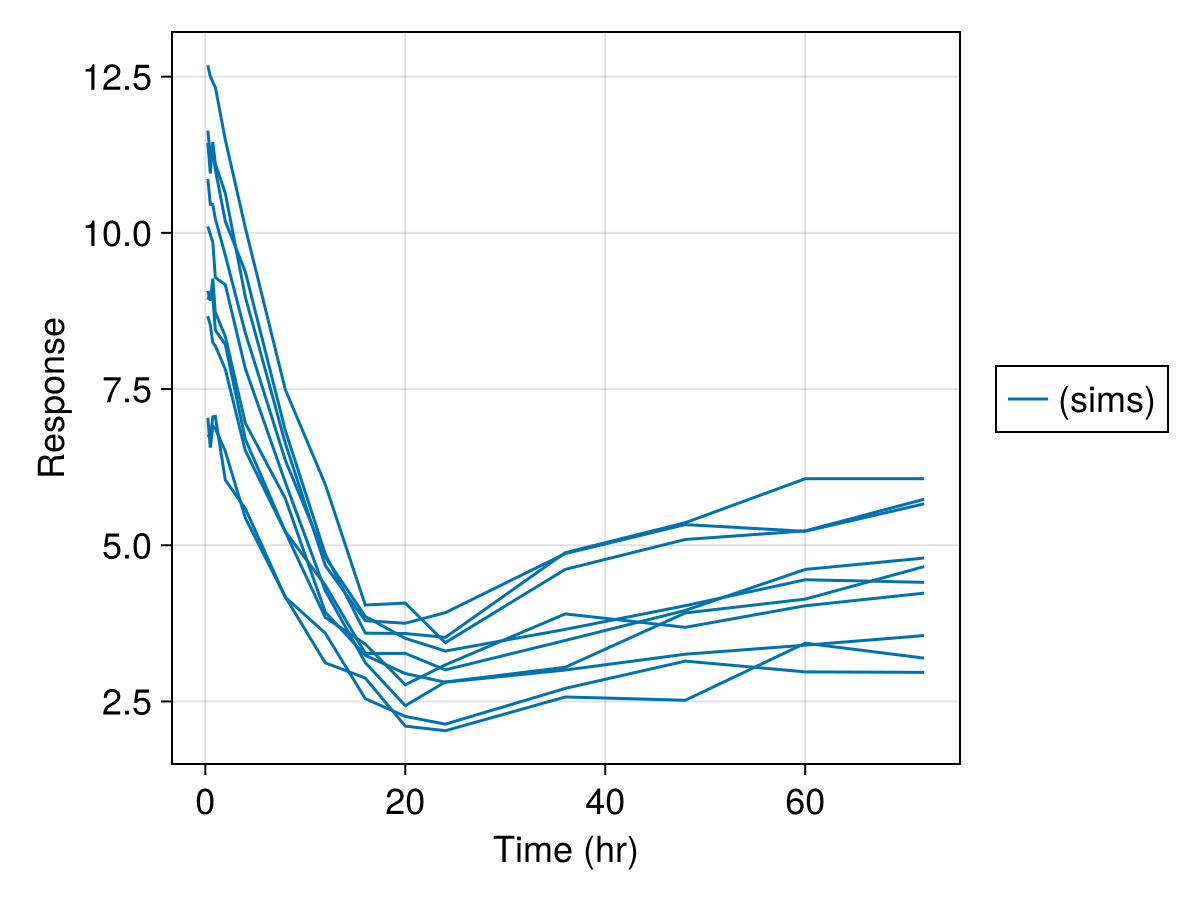
sim_plot(
inf_2cmt_lin_turnover,
sims;
observations = [:resp],
figure = (; fontsize = 18),
axis = (; xlabel = "Time (hr)", ylabel = "Response"),
)
In this code, we defined a nonlinear mixed effects model by describing the parameters, the random effects, the dynamical model, and the derived (result) values. Then we generated a population of 10 subjects who received a single dose of 150 mg, specified parameter values, simulated the model, and generated a plot of the results. Now let's walk through this process!
Using the Model Macro
First we define the model. The simplest way to do this is via @model. Inside the @model block we have a few subsections. The first of these is a @param block.
Pumas.@param — Macro@paramDefines the model's fixed effects and corresponding domains, e.g. tvcl ∈ RealDomain(lower = 0). Must be used in an @model block. For example:
@model begin
@param begin
tvcl ∈ RealDomain(lower = 0)
tvv ∈ RealDomain(lower = 0)
Ω ∈ PDiagDomain(2)
σ_prop ∈ RealDomain(lower = 0, init = 0.04)
end
endFor PumasEMModels, the @param block uses a formula syntax for covariate relationships. A distribution from the normal family is used to indicate domain (currently only Normal, LogNormal, and LogitNormal are supported):
@emmodel begin
@param begin
CL ~ 1 + wt | LogNormal
Vc ~ 1 | Normal
end
endthis gives CL = exp(log(θ.CL[1]) + θ.CL[2] * wt) and VC = θ.VC, where θ is the parameter named tuple. If we only have 1 in the formula, then it is a scalar. If we have covariates, we have a tuple of parameters for variable.
For this model we define structural model parameters of PK and PD and their corresponding variances where applicable:
@param begin
tvcl ∈ RealDomain(; lower = 0)
tvvc ∈ RealDomain(; lower = 0)
tvq ∈ RealDomain(; lower = 0)
tvvp ∈ RealDomain(; lower = 0)
Ω_pk ∈ PDiagDomain(4)
σ_prop_pk ∈ RealDomain(; lower = 0)
σ_add_pk ∈ RealDomain(; lower = 0)
# PD parameters
tvturn ∈ RealDomain(; lower = 0)
tvebase ∈ RealDomain(; lower = 0)
tvec50 ∈ RealDomain(; lower = 0)
Ω_pd ∈ PDiagDomain(1)
σ_add_pd ∈ RealDomain(; lower = 0)
endNote the mathematical elegance of the expression of parameters, relationships, and distributions.
Next we define our random effects using the @random block.
Pumas.@random — Macro@randomdefines the model's random effects and corresponding distribution, e.g. η ~ MvNormal(Ω). Must be used in an @model block. For example:
@model begin
@random begin
η ~ MvNormal(Ω)
end
endFor PumasEMModels, the @random block is equivalent to the @params block, except that each variable defined here has an additional random effect. For example:
@emmodel begin
@random begin
CL ~ 1 + wt | LogNormal
Vc ~ 1 | Normal
end
endthis gives CL = exp(log(θ.CL[1]) + θ.CL[2] * wt + η_CL) and VC = θ.VC + η_VC, where θ is the parameter named tuple. If we only have 1 in the formula, then it is a scalar. If we have covariates, we have a tuple of parameters for variable.
The random effects are defined by a distribution from the Distributions package. For more information on defining distributions, please see the Distributions documentation. For this tutorial, we wish to have a multivariate normal of uncorrelated random effects, one for PK and another for PD, so we utilize the syntax:
@random begin
ηpk ~ MvNormal(Ω_pk)
ηpd ~ MvNormal(Ω_pd)
endNow we define our pre-processing step in @pre.
Pumas.@pre — Macro@prePre-processes variables for the dynamic system and statistical specification. Must be used in an @model block. For example:
@model begin
@params begin
tvcl ∈ RealDomain(lower = 0)
tvv ∈ RealDomain(lower = 0)
ωCL ∈ RealDomain(lower = 0)
ωV ∈ RealDomain(lower = 0)
σ_prop ∈ RealDomain(lower = 0, init = 0.04)
end
@random begin
ηCL ~ Normal(ωCL)
ηV ~ Normal(ωV)
end
@covariates wt
@pre begin
CL = tvcl * (wt / 70)^0.75 * exp(ηCL)
Vc = tvv * (wt / 70) * exp(ηV)
ka = CL / Vc
Q = Vc
end
endFor our PK/PD model, we define the individual parameters and their distributions, and we give them names as follows:
@pre begin
CL = tvcl * exp(ηpk[1])
Vc = tvvc * exp(ηpk[2])
Q = tvq * exp(ηpk[3])
Vp = tvvp * exp(ηpk[4])
ebase = tvebase * exp(ηpd[1])
ec50 = tvec50
emax = 1
turn = tvturn
kout = 1 / turn
kin0 = ebase * kout
endNext we define the @init block which specifies the initial conditions of the dynamic system (i.e., initial compartment amounts).
Pumas.@init — Macro@initDefines initial conditions of the dynamic system. Must be used in an @model block. For example:
@model begin
@init begin
Depot1 = 0.0
Depot2 = 0.0
Central = 0.0
end
endAny variable not mentioned in this block is assumed to have a zero for its starting value. We wish to only set the starting value for Resp, and thus we use:
@init begin
Resp = ebase
endNow we define our dynamics. We do this via the @dynamics block.
Pumas.@dynamics — Macro@dynamicsDefines the dynamic system by specifying differential equations. Must be used in an @model block.
A @model block with a @dynamics block must not contain a @reactions block.
Examples
A @dynamics block with a system of differential equations:
@model begin
@dynamics begin
Depot1' = -ka * Depot1
Depot2' = ka * Depot1 - ka * Depot2
Central' = ka * Depot2 - CL / Vc * Central
end
endA @dynamics block with a differential equation model:
@model begin
@dynamics Depots1Central1
endFor our PK/PD model, we specify the three differential equations that describe the PK and PD components of the system:
@dynamics begin
Central' = -(CL / Vc) * Central + (Q / Vp) * Peripheral - (Q / Vc) * Central
Peripheral' = (Q / Vc) * Central - (Q / Vp) * Peripheral
Resp' = kin - kout * Resp
endNote the convenience of expressing the compartment amounts by name and the value of the derivative equations using the single quote character.
Next we set up alias variables that can be used later in the code. Such alias code can be setup in the @vars block.
Pumas.@vars — Macro@varsDefine variables usable in other blocks. It is recommended to define them in @pre instead if not a function of dynamic variables. Must be used in an @model block. For example:
@model begin
@vars begin
conc = Central / Vc
end
endWe create aliases for the concentration and response as follows:
@vars begin
conc := Central / Vc
edrug := emax * conc / (ec50 + conc)
kin := kin0 * (1 - edrug)
endNote that the local assignment := can be used to define intermediate statements that will not be carried outside the block. This means that all the resulting data workflows from this model will not contain the intermediate variables defined with :=. This optional use of local assignment improves computational efficiency.
Note that without adjustment the units of conc are the units of the dose divided by the units of the volume parameter. The balancing of conc units with those reported from the assay could occur here, but in this example we make this adjustment below in the @derived block.
Lastly we utilize the @derived block.
Pumas.@derived — Macro@derivedDefines the error model of the dependent variables. Must be used in an @model block. For example:
@model begin
@derived begin
dv ~ @. Normal(1000 * Central / Vc, 1000 * Central / Vc * σ)
end
endOur error model includes a component for each type of observation (i.e., conc and Resp):
@derived begin
dv ~ @. Normal(conc, sqrt((conc * σ_prop_pk)^2 + σ_add_pk^2))
resp ~ @. Normal(Resp, σ_add_pd)
endHere dv (i.e., dependent variable) is the drug concentration and resp is the response. At any time that these outputs are sampled dv is normally distributed with mean conc and variance (conc * σ_prop_pk)^2 + σ_add_pk^2, and resp has mean Resp and a constant standard deviation σ_add_pd.
Building a Subject
Now let's build a subject to simulate the model with.
Pumas.Subject — TypeSubjectThe data corresponding to a single subject:
Fields:
id: identifierobservations: a named tuple of the dependent variablescovariates: a named tuple containing the covariates, ornothing.events: aDosageRegimen, ornothing.time: a vector of time stamps for the observations
When there are time varying covariates, each covariate is interpolated with a common covariate time support. The interpolated values are then used to build a multi-valued interpolant for the complete time support.
From the multi-valued interpolant, certain discontinuities are flagged in order to use that information for the differential equation solvers and to correctly apply the analytical solution per region as applicable.
Constructor
Subject(;id = "1",
observations = NamedTuple(),
events = nothing,
time = observations isa AbstractDataFrame ? observations.time : nothing,
covariates::Union{Nothing, NamedTuple} = nothing,
covariates_time = observations isa AbstractDataFrame ? observations.time : nothing,
covariates_direction = :left)Subject may be constructed from an <:AbstractDataFrame with the appropriate schema or by providing the arguments directly through separate DataFrames / structures.
Examples:
julia> Subject()
Subject
ID: 1
julia> Subject(
id = 20,
events = DosageRegimen(200, ii = 24, addl = 2),
covariates = (WT = 14.2, HT = 5.2),
)
Subject
ID: 20
Events: 3
Covariates: WT, HT
julia> Subject(covariates = (WT = [14.2, 14.7], HT = fill(5.2, 2)), covariates_time = [0, 3])
Subject
ID: 1
Covariates: WT, HTOur model did not make use of covariates, so we will ignore covariate components covariates, covariate_time and covariate_direction for now, and observations are only necessary for fitting parameters to data which will not be covered in this tutorial. Thus, our subject will be defined simply by its dosage regimen.
To do this, we use the DosageRegimen constructor.
Pumas.DosageRegimen — TypeDosageRegimen(
amt::Numeric;
time::Numeric = 0,
cmt::Union{Numeric,Symbol} = 1,
evid::Numeric = 1,
ii::Numeric = zero.(time),
addl::Numeric = 0,
rate::Numeric = zero.(amt)./oneunit.(time),
duration::Numeric = zero(amt)./oneunit.(time),
ss::Numeric = 0,
route::NCA.Route = NCA.NullRoute,
)Lazy representation of a series of events.
Examples
julia> DosageRegimen(100; ii = 24, addl = 6)
DosageRegimen
Row │ time cmt amt evid ii addl rate duration ss route
│ Float64 Int64 Float64 Int8 Float64 Int64 Float64 Float64 Int8 NCA.Route
─────┼───────────────────────────────────────────────────────────────────────────────────
1 │ 0.0 1 100.0 1 24.0 6 0.0 0.0 0 NullRoute
julia> DosageRegimen(50; ii = 12, addl = 13)
DosageRegimen
Row │ time cmt amt evid ii addl rate duration ss route
│ Float64 Int64 Float64 Int8 Float64 Int64 Float64 Float64 Int8 NCA.Route
─────┼───────────────────────────────────────────────────────────────────────────────────
1 │ 0.0 1 50.0 1 12.0 13 0.0 0.0 0 NullRoute
julia> DosageRegimen(200; ii = 24, addl = 2)
DosageRegimen
Row │ time cmt amt evid ii addl rate duration ss route
│ Float64 Int64 Float64 Int8 Float64 Int64 Float64 Float64 Int8 NCA.Route
─────┼───────────────────────────────────────────────────────────────────────────────────
1 │ 0.0 1 200.0 1 24.0 2 0.0 0.0 0 NullRoute
julia> DosageRegimen(200; ii = 24, addl = 2, route = NCA.IVBolus)
DosageRegimen
Row │ time cmt amt evid ii addl rate duration ss route
│ Float64 Int64 Float64 Int8 Float64 Int64 Float64 Float64 Int8 NCA.Route
─────┼───────────────────────────────────────────────────────────────────────────────────
1 │ 0.0 1 200.0 1 24.0 2 0.0 0.0 0 IVBolusYou can also create a new DosageRegimen from various existing DosageRegimens:
evs = DosageRegimen(
regimen1::DosageRegimen,
regimen2::DosageRegimen;
offset = nothing
)offset specifies if regimen2 should start after an offset following the end of the last event in regimen1.
Examples
julia> e1 = DosageRegimen(100; ii = 24, addl = 6)
DosageRegimen
Row │ time cmt amt evid ii addl rate duration ss route
│ Float64 Int64 Float64 Int8 Float64 Int64 Float64 Float64 Int8 NCA.Route
─────┼───────────────────────────────────────────────────────────────────────────────────
1 │ 0.0 1 100.0 1 24.0 6 0.0 0.0 0 NullRoute
julia> e2 = DosageRegimen(50; ii = 12, addl = 13)
DosageRegimen
Row │ time cmt amt evid ii addl rate duration ss route
│ Float64 Int64 Float64 Int8 Float64 Int64 Float64 Float64 Int8 NCA.Route
─────┼───────────────────────────────────────────────────────────────────────────────────
1 │ 0.0 1 50.0 1 12.0 13 0.0 0.0 0 NullRoute
julia> evs = DosageRegimen(e1, e2)
DosageRegimen
Row │ time cmt amt evid ii addl rate duration ss route
│ Float64 Int64 Float64 Int8 Float64 Int64 Float64 Float64 Int8 NCA.Route
─────┼───────────────────────────────────────────────────────────────────────────────────
1 │ 0.0 1 100.0 1 24.0 6 0.0 0.0 0 NullRoute
2 │ 0.0 1 50.0 1 12.0 13 0.0 0.0 0 NullRoute
julia> DosageRegimen(e1, e2; offset = 10)
DosageRegimen
Row │ time cmt amt evid ii addl rate duration ss route
│ Float64 Int64 Float64 Int8 Float64 Int64 Float64 Float64 Int8 NCA.Route
─────┼───────────────────────────────────────────────────────────────────────────────────
1 │ 0.0 1 100.0 1 24.0 6 0.0 0.0 0 NullRoute
2 │ 178.0 1 50.0 1 12.0 13 0.0 0.0 0 NullRouteFor our example, let's assume the dose is a 150 mg infusion administered at the rate of 10 mg/hour into the Central compartment:
regimen = DosageRegimen(150; time = 0, rate = 10, cmt = :Central)| Row | time | cmt | amt | evid | ii | addl | rate | duration | ss | route |
|---|---|---|---|---|---|---|---|---|---|---|
| Float64 | Symbol | Float64 | Int8 | Float64 | Int64 | Float64 | Float64 | Int8 | NCA.Route | |
| 1 | 0.0 | Central | 150.0 | 1 | 0.0 | 0 | 10.0 | 15.0 | 0 | NullRoute |
Let's define our subject to have id=1 with this single-dose regimen:
subject = Subject(; id = 1, events = regimen)Subject
ID: 1
Events: 1We can also create a collection of subjects which forms a population.
pop = map(i -> Subject(; id = i, events = regimen), 1:10)Population
Subjects: 10
Observations: Running a Simulation
The main function for running a simulation is simobs.
Pumas.simobs — Functionsimobs(
model::AbstractPumasModel,
population::Union{Subject, Population},
param,
randeffs=sample_randeffs(model, param, population);
obstimes=nothing,
ensemblealg=EnsembleSerial(),
diffeq_options=NamedTuple(),
rng=Random.default_rng(),
simulate_error=Val(true),
)Simulate random observations from model for population with parameters param at obstimes (by default, use the times of the existing observations for each subject). If no randeffs is provided, then random ones are generated according to the distribution in the model.
Arguments
modelmay either be aPumasModelor aPumasEMModel.populationmay either be aPopulationofSubjects or a singleSubject.paramshould be either a single parameter set, in the form of aNamedTuple, or a vector of such parameter sets. If a vector then each of the parameter sets in that vector will be applied in turn. Example:(; tvCL=1., tvKa=2., tvV=1.)randeffsis an optional argument that, if used, should specify the random effects for each subject inpopulation. Such random effects are specified byNamedTuples forPumasModels(e.g.(; tvCL=1., tvKa=2.)) and byTuplesforPumasEMModels (e.g.(1., 2.)). Ifpopulationis a singleSubject(without being enclosed in a vector) then a single such random effect specifier should be passed. If, however,populationis aPopulationof multipleSubjects thenrandeffsshould be a vector containing one such specifier for eachSubject. The functionsinit_randeffs,zero_randeffs, andsample_randeffsare sometimes convenient for generatingrandeffs:
randeffs = zero_randeffs(model, param, population)
solve(model, population, param, randeffs)If no randeffs is provided, then random ones are generated according to the distribution in the model.
obstimesis a keyword argument where you can pass a vector of times at which to simulate observations. The default,nothing, ensures the use of the existing observation times for eachSubject.ensemblealgis a keyword argument that allows you to choose between different modes of parallelization. Options includeEnsembleSerial(),EnsembleThreads()andEnsembleDistributed().diffeq_optionsis a keyword argument that takes aNamedTupleof options to pass on to the differential equation solver.rngis a keyword argument where you can specify which random number generator to use.simulate_erroris a keyword argument that allows you to choose whether a sample (Val(true)ortrue) or the mean (Val(false)orfalse) of the predictive distribution's error model will be returned for each subject.
simobs(model::PumasModel, population::Population; samples::Int = 10, simulate_error = true, rng::AbstractRNG = default_rng())Simulates model predictions for population using the prior distributions for the population and subject-specific parameters. samples is the number of simulated predictions returned per subject. If simulate_error is false, the mean of the predictive distribution's error model will be returned, otherwise a sample from the predictive distribution's error model will be returned for each subject. rng is the pseudo-random number generator used in the sampling.
simobs(model::PumasModel, subject::Subject; samples::Int = 10, simulate_error = true, rng::AbstractRNG = default_rng())Simulates model predictions for subject using the prior distributions for the population and subject-specific parameters. samples is the number of simulated predictions returned. If simulate_error is false, the mean of the predictive distribution's error model will be returned, otherwise a sample from the predictive distribution's error model will be returned for each subject. rng is the pseudo-random number generator used in the sampling.
simobs(b::BayesMCMCResults; subject = nothing, samples::Int = 10, simulate_error = false, rng::AbstractRNG = default_rng())Simulates model predictions using the posterior samples for the population and subject-specific parameters. The predictions are either for the subject with index subject or the whole population if no index is input. samples is the number of simulated predictions returned per subject. If simulate_error is false, the mean of the predictive distribution's error model will be returned, otherwise a sample from the predictive distribution's error model will be returned for each subject. rng is the pseudo-random number generator used in the sampling.
simobs(pmi::FittedPumasModelInference, population::Population = pmi.fpm.data, randeffs::Union{Nothing, AbstractVector{<:NamedTuple}} = nothing; samples::Int, rng = default_rng(), kwargs...,)Simulates observations from the fitted model using a truncated multivariate normal distribution for the parameter values when a covariance estimate is available and using non-parametric sampling for simulated inference and bootstrapping. When a covariance estimate is available, the optimal parameter values are used as the mean and the variance-covariance estimate is used as the covariance matrix. Rejection sampling is used to avoid parameter values outside the parameter domains. Each sample uses a different parameter value. samples is the number of samples to sample. population is the population of subjects which defaults to the population associated the fitted model. randeffs can be set to a vector of named tuples, one for each sample. If randeffs is not specified (the default behaviour), it will be sampled from its distribution.
simobs(fpm::FittedPumasModel, [population::Population,] vcov::AbstractMatrix, randeffs::Union{Nothing, AbstractVector{<:NamedTuple}} = nothing; samples::Int, rng = default_rng(), kwargs...)Simulates observations from the fitted model using a truncated multi-variate normal distribution for the parameter values. The optimal parameter values are used for the mean and the user supplied variance-covariance (vcov) is used as the covariance matrix. Rejection sampling is used to avoid parameter values outside the parameter domains. Each sample uses a different parameter value. samples is the number of samples to sample. population is the population of subjects which defaults to the population associated the fitted model so it's optional to pass. randeffs can be set to a vector of named tuples, one for each sample. If randeffs is not specified (the default behaviour), it will be sampled from its distribution.
Let's simulate subject 1 with a set of chosen parameters:
turnover_params = (;
tvcl = 1.5,
tvvc = 25.0,
tvq = 5.0,
tvvp = 150.0,
tvturn = 10,
tvebase = 10,
tvec50 = 0.3,
Ω_pk = Diagonal([0.05, 0.05, 0.05, 0.05]),
Ω_pd = Diagonal([0.05]),
σ_prop_pk = 0.15,
σ_add_pk = 0.5,
σ_add_pd = 0.2,
)
sims = simobs(inf_2cmt_lin_turnover, subject, turnover_params; obstimes = sd_obstimes)SimulatedObservations
Simulated variables: dv, resp
Time: [0.25, 0.5, 0.75, 1.0, 2.0, 4.0, 8.0, 12.0, 16.0, 20.0, 24.0, 36.0, 48.0, 60.0, 71.9]We can then plot the simulated observations by using the sim_plot function:
sim_plot(inf_2cmt_lin_turnover, sims; observations = :dv)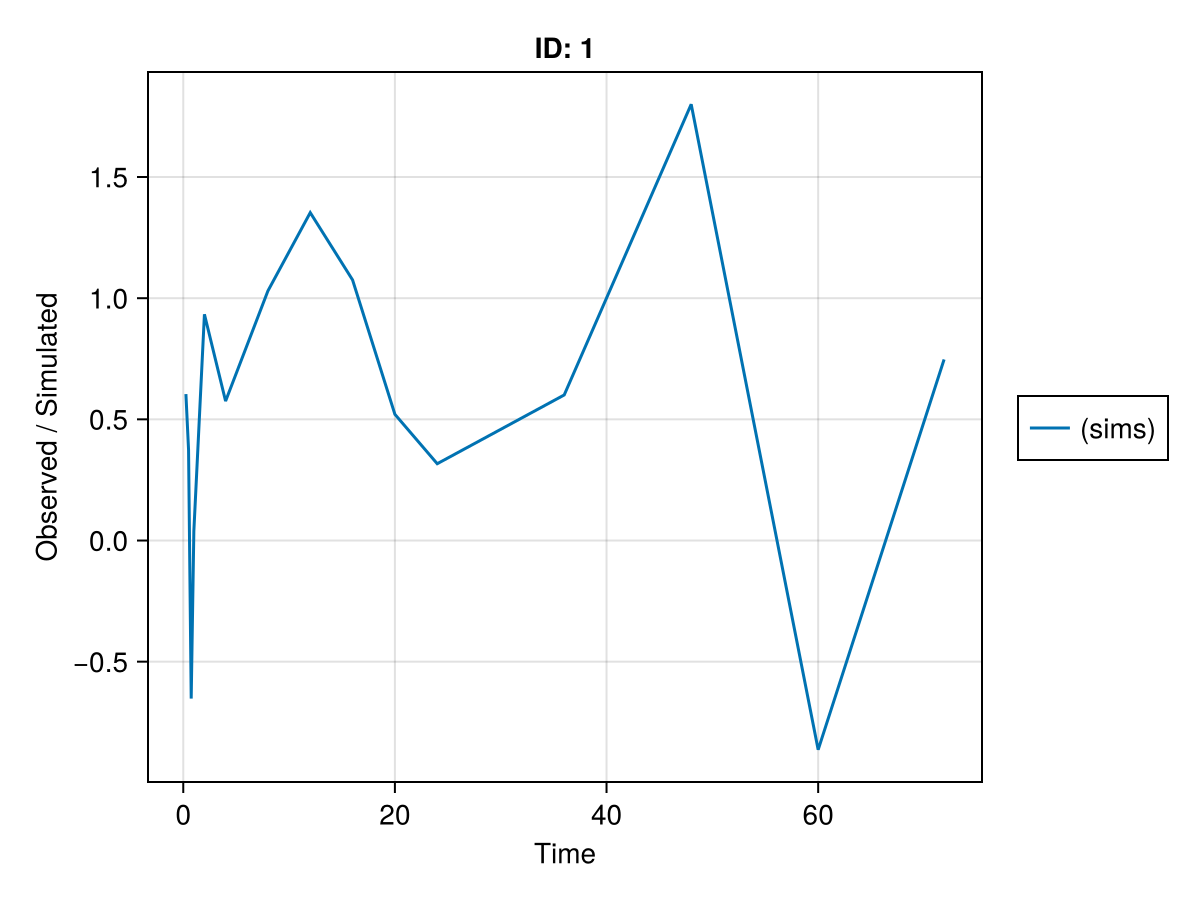
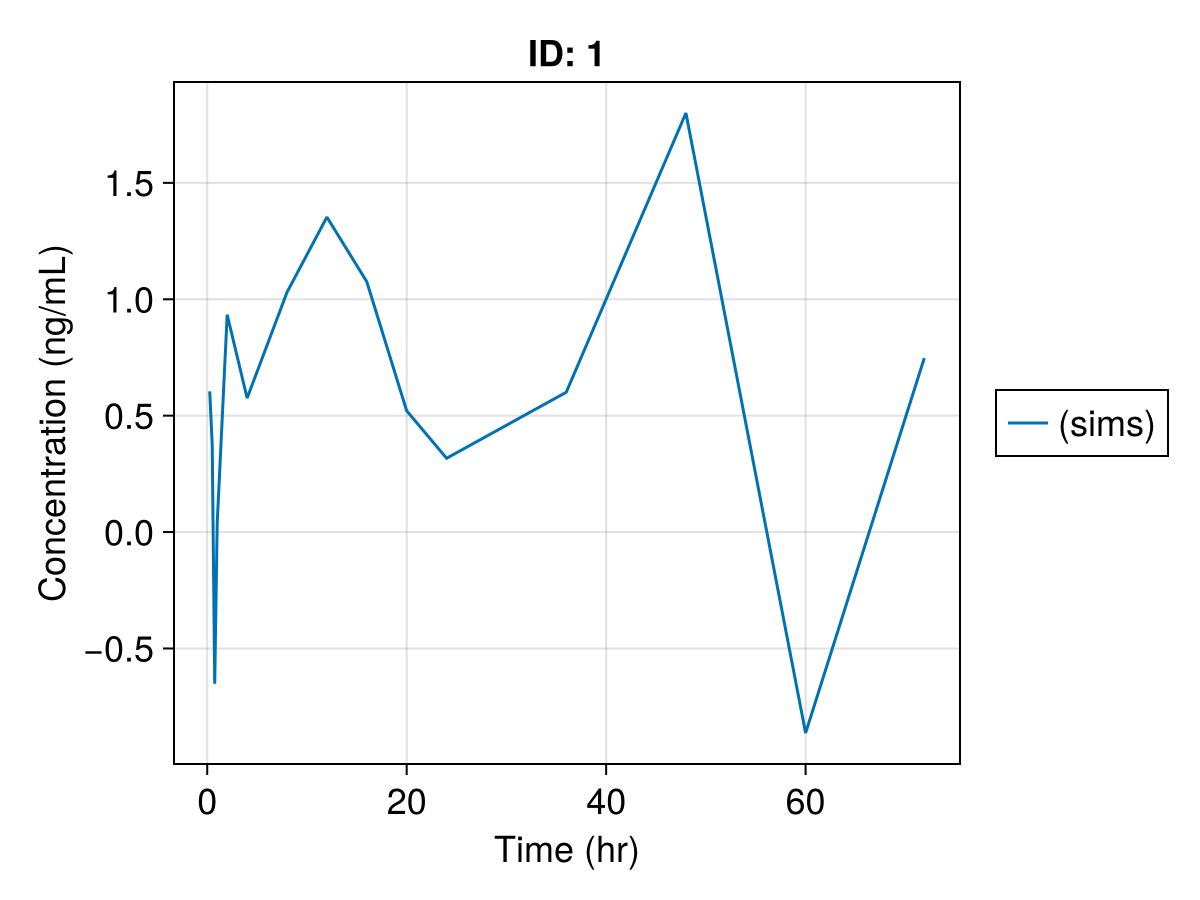
Note that we can further modify the plot, see Customizing Plots. For example,
sim_plot(
inf_2cmt_lin_turnover,
sims,
observations = :dv,
figure = (; fontsize = 18),
axis = (; xlabel = "Time (hr)", ylabel = "Concentration (ng/mL)"),
)
When we only give the structural model parameters, the random effects are automatically sampled from their distributions. If we wish to prescribe a value for the random effects, we pass initial values similarly:
vrandeffs = [(; ηpk = rand(4), ηpd = rand()) for _ in pop]
sims =
simobs(inf_2cmt_lin_turnover, pop, turnover_params, vrandeffs; obstimes = sd_obstimes)Simulated population (Vector{<:Subject})
Simulated subjects: 10
Simulated variables: dv, respIf a population simulation is required with the inter-subject random effects set to zero, then it is possible to use the zero_randeffs function:
etas = zero_randeffs(inf_2cmt_lin_turnover, turnover_params, pop)
sims = simobs(inf_2cmt_lin_turnover, pop, turnover_params, etas; obstimes = sd_obstimes)
fig = Figure(; resolution = (1000, 600), fontsize = 18)
ax1 = Axis(fig[1, 1]; xlabel = "Time (hr)", ylabel = "Concentration (ng/mL)")
ax2 = Axis(fig[1, 2]; xlabel = "Time (hr)", ylabel = "Response")
sim_plot!(ax1, inf_2cmt_lin_turnover, sims; observations = [:dv])
sim_plot!(ax2, inf_2cmt_lin_turnover, sims; observations = [:resp])
fig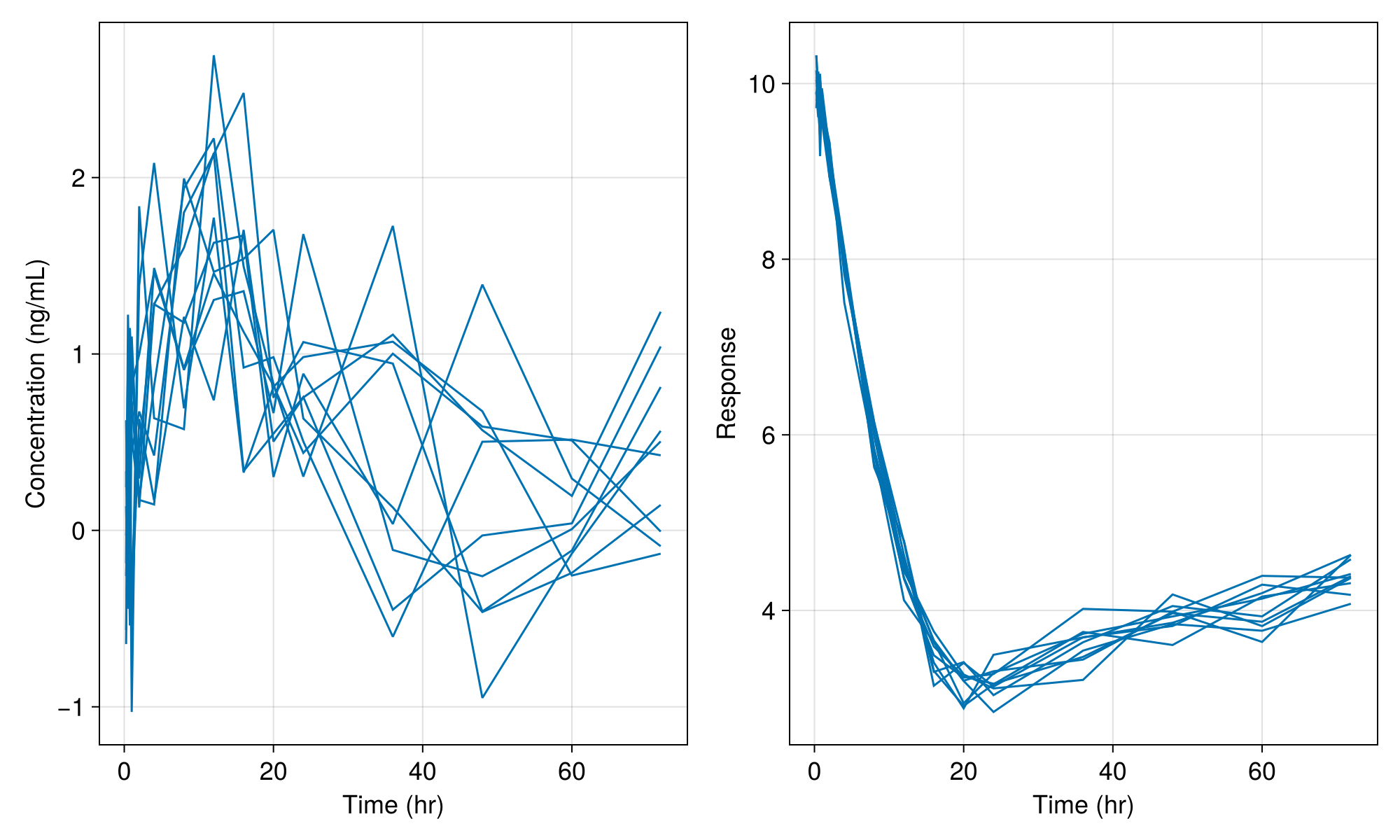
We still see variability in the plot above due to the residual variability components in the model. If we set the residual error variance parameters to tiny values as below
turnover_params_wo_sigma =
(turnover_params..., σ_prop_pk = 1e-20, σ_add_pk = 1e-20, σ_add_pd = 1e-20)(tvcl = 1.5,
tvvc = 25.0,
tvq = 5.0,
tvvp = 150.0,
tvturn = 10,
tvebase = 10,
tvec50 = 0.3,
Ω_pk = [0.05 0.0 0.0 0.0; 0.0 0.05 0.0 0.0; 0.0 0.0 0.05 0.0; 0.0 0.0 0.0 0.05],
Ω_pd = [0.05;;],
σ_prop_pk = 1.0e-20,
σ_add_pk = 1.0e-20,
σ_add_pd = 1.0e-20,)and perform the simulation without residual random effects we get only the population mean:
etas = zero_randeffs(inf_2cmt_lin_turnover, turnover_params, pop)
sims = simobs(
inf_2cmt_lin_turnover,
pop,
turnover_params_wo_sigma,
etas;
obstimes = sd_obstimes,
)
fig = Figure(; resolution = (1000, 600), fontsize = 18)
ax1 = Axis(fig[1, 1]; xlabel = "Time (hr)", ylabel = "Concentration (ng/mL)")
ax2 = Axis(fig[1, 2]; xlabel = "Time (hr)", ylabel = "Response")
sim_plot!(ax1, inf_2cmt_lin_turnover, sims; observations = [:dv])
sim_plot!(ax2, inf_2cmt_lin_turnover, sims; observations = [:resp])
fig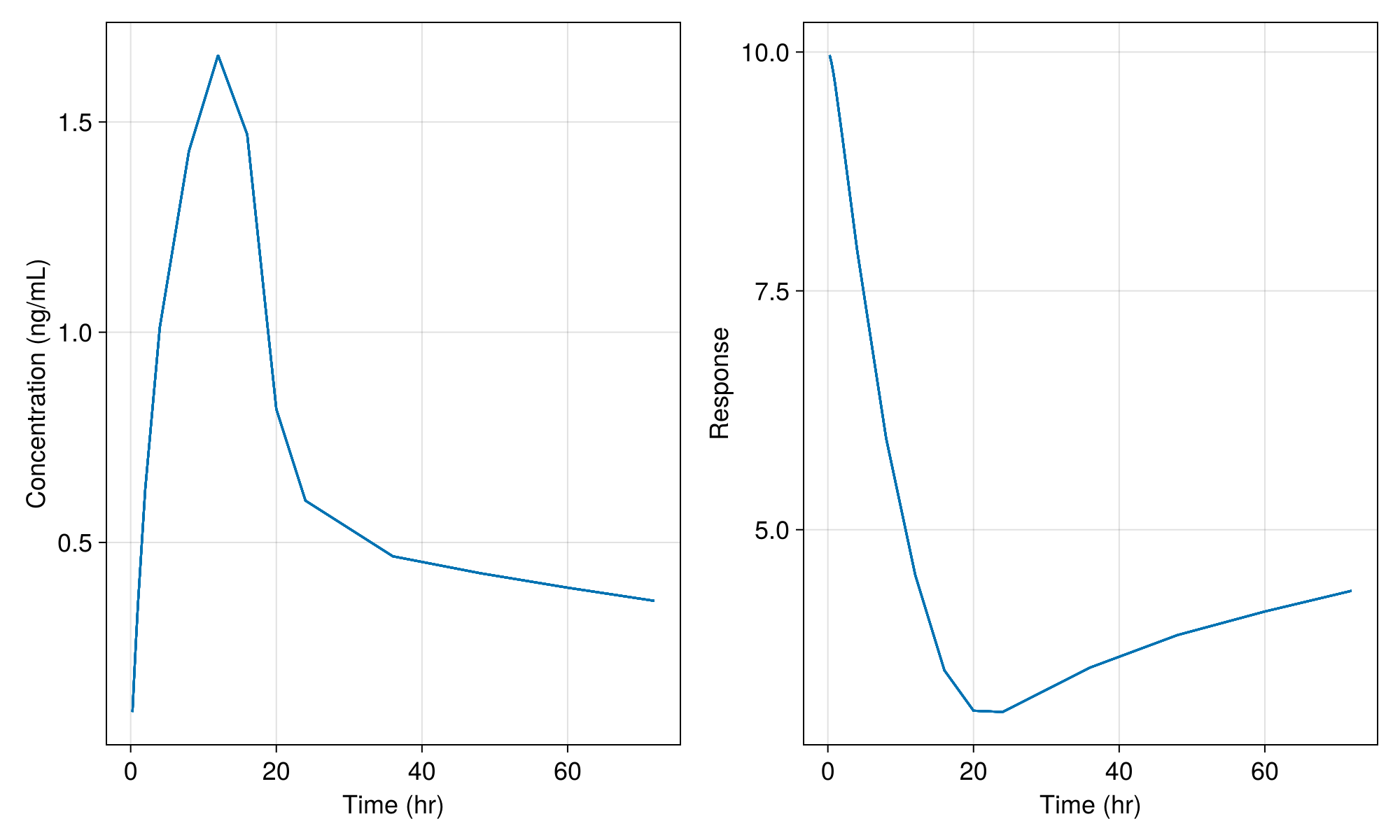
Handling the resulting simulation data
The resulting sims object contains the results of the simulation for each subject. The results for the ith subject can be retrieved by sims[i].
We can convert the simulation results to Pumas Subjects using the constructor broadcasted over all elements of sims:
sims_subjects = Subject.(sims)Population
Subjects: 10
Observations: dv, respThe newly created Subjects will contain the variables defined in the @derived-block as their observations. Then sims_subjects can be used to fit the model parameters based on the simulated dataset. We can also turn the simulated population into a DataFrame with the DataFrame constructor:
DataFrame(sims[1])| Row | id | time | dv | resp | evid | amt | cmt | rate | duration | ss | ii | route | Central | Peripheral | Resp | CL | Vc | Q | Vp | ebase | ec50 | emax | turn | kout | kin0 | ηpk_1 | ηpk_2 | ηpk_3 | ηpk_4 | ηpd_1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| String | Float64 | Float64? | Float64? | Int64 | Float64? | Symbol? | Float64? | Float64? | Int8? | Float64? | NCA.Route? | Float64? | Float64? | Float64? | Float64? | Float64? | Float64? | Float64? | Float64? | Float64? | Int64? | Int64? | Float64? | Float64? | Float64 | Float64 | Float64 | Float64 | Float64 | |
| 1 | 1 | 0.0 | missing | missing | 1 | 150.0 | Central | 10.0 | 15.0 | 0 | 0.0 | NullRoute | missing | missing | missing | 1.5 | 25.0 | 5.0 | 150.0 | 10.0 | 0.3 | 1 | 10 | 0.1 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 2 | 1 | 0.25 | 0.096826 | 9.96663 | 0 | missing | missing | missing | missing | missing | missing | missing | 2.42065 | 0.0609992 | 9.96663 | 1.5 | 25.0 | 5.0 | 150.0 | 10.0 | 0.3 | 1 | 10 | 0.1 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 3 | 1 | 0.5 | 0.187597 | 9.88839 | 0 | missing | missing | missing | missing | missing | missing | missing | 4.68993 | 0.238203 | 9.88839 | 1.5 | 25.0 | 5.0 | 150.0 | 10.0 | 0.3 | 1 | 10 | 0.1 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 4 | 1 | 0.75 | 0.272731 | 9.78409 | 0 | missing | missing | missing | missing | missing | missing | missing | 6.81828 | 0.523374 | 9.78409 | 1.5 | 25.0 | 5.0 | 150.0 | 10.0 | 0.3 | 1 | 10 | 0.1 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 5 | 1 | 1.0 | 0.352616 | 9.66347 | 0 | missing | missing | missing | missing | missing | missing | missing | 8.8154 | 0.908843 | 9.66347 | 1.5 | 25.0 | 5.0 | 150.0 | 10.0 | 0.3 | 1 | 10 | 0.1 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 6 | 1 | 2.0 | 0.626531 | 9.10546 | 0 | missing | missing | missing | missing | missing | missing | missing | 15.6633 | 3.31813 | 9.10546 | 1.5 | 25.0 | 5.0 | 150.0 | 10.0 | 0.3 | 1 | 10 | 0.1 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 7 | 1 | 4.0 | 1.01152 | 7.93649 | 0 | missing | missing | missing | missing | missing | missing | missing | 25.2881 | 11.1916 | 7.93649 | 1.5 | 25.0 | 5.0 | 150.0 | 10.0 | 0.3 | 1 | 10 | 0.1 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 8 | 1 | 8.0 | 1.43067 | 5.95748 | 0 | missing | missing | missing | missing | missing | missing | missing | 35.7668 | 33.2305 | 5.95748 | 1.5 | 25.0 | 5.0 | 150.0 | 10.0 | 0.3 | 1 | 10 | 0.1 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 9 | 1 | 12.0 | 1.65869 | 4.52562 | 0 | missing | missing | missing | missing | missing | missing | missing | 41.4673 | 58.211 | 4.52562 | 1.5 | 25.0 | 5.0 | 150.0 | 10.0 | 0.3 | 1 | 10 | 0.1 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 10 | 1 | 16.0 | 1.47042 | 3.52807 | 0 | missing | missing | missing | missing | missing | missing | missing | 36.7604 | 82.7287 | 3.52807 | 1.5 | 25.0 | 5.0 | 150.0 | 10.0 | 0.3 | 1 | 10 | 0.1 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 11 | 1 | 20.0 | 0.81701 | 3.10522 | 0 | missing | missing | missing | missing | missing | missing | missing | 20.4252 | 92.5612 | 3.10522 | 1.5 | 25.0 | 5.0 | 150.0 | 10.0 | 0.3 | 1 | 10 | 0.1 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 12 | 1 | 24.0 | 0.599389 | 3.09228 | 0 | missing | missing | missing | missing | missing | missing | missing | 14.9847 | 93.8667 | 3.09228 | 1.5 | 25.0 | 5.0 | 150.0 | 10.0 | 0.3 | 1 | 10 | 0.1 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 13 | 1 | 36.0 | 0.467029 | 3.55706 | 0 | missing | missing | missing | missing | missing | missing | missing | 11.6757 | 87.9834 | 3.55706 | 1.5 | 25.0 | 5.0 | 150.0 | 10.0 | 0.3 | 1 | 10 | 0.1 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 14 | 1 | 48.0 | 0.426772 | 3.89715 | 0 | missing | missing | missing | missing | missing | missing | missing | 10.6693 | 80.9633 | 3.89715 | 1.5 | 25.0 | 5.0 | 150.0 | 10.0 | 0.3 | 1 | 10 | 0.1 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 15 | 1 | 60.0 | 0.392367 | 4.14396 | 0 | missing | missing | missing | missing | missing | missing | missing | 9.80916 | 74.4559 | 4.14396 | 1.5 | 25.0 | 5.0 | 150.0 | 10.0 | 0.3 | 1 | 10 | 0.1 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 16 | 1 | 71.9 | 0.361073 | 4.36054 | 0 | missing | missing | missing | missing | missing | missing | missing | 9.02683 | 68.5177 | 4.36054 | 1.5 | 25.0 | 5.0 | 150.0 | 10.0 | 0.3 | 1 | 10 | 0.1 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
From there, any Julia tools can be used to analyze these arrays and DataFrames. Or you can conveniently export this to a CSV, Excel, or SAS data file.
Conducting an Estimation
With a model and population you can fit your model to your data. Model fitting in Pumas has the purpose of estimating parameters of a model, given a dataset, and is performed by calling the fit function with the following positional arguments:
- a
PumasModelconstructed via@model, or aPumasEMModelconstructed via@emmodel. - a
Populationor aSubject. - a named tuple of the initial parameter values.
- an inference algorithm.
If you want to use the model's initial parameter values declared inside the @param block, you can do so with init_params(model). Note that this will fall back to the parameters' default values if you do not specify an initial value inside the @param block.
Pumas supports the following inference algorithms:
Maximum Likelihood Estimation:
NaivePooled: first order approximation without inter-subject random-effects.FO: first-order approximation.FOCE: first-order conditional estimation.LaplaceI: second-order Laplace approximation.SAEM: Stochastic Approximation using the Expectation-Maximization algorithm. Note that for this you use@emmodelinstead of@model.
Bayesian inference:
BayesMCMC: MCMC using No-U-Turn Sampler (NUTS).MarginalMCMC: MCMC using No-U-Turn Sampler (NUTS), but marginalizing out the subject-specific parameters.MAP: Maximum A Posteriori estimation.
Now we can simultaneously fit our PK & PD models using one of the simulated populations from the previous section:
turnover_params = (;
tvcl = 1.5,
tvvc = 25.0,
tvq = 5.0,
tvvp = 150.0,
tvturn = 10,
tvebase = 10,
tvec50 = 0.3,
Ω_pk = Diagonal([0.05, 0.05, 0.05, 0.05]),
Ω_pd = Diagonal([0.05]),
σ_prop_pk = 0.15,
σ_add_pk = 0.5,
σ_add_pd = 0.2,
)
sims = simobs(inf_2cmt_lin_turnover, pop, turnover_params; obstimes = sd_obstimes)Simulated population (Vector{<:Subject})
Simulated subjects: 10
Simulated variables: dv, respfit_2cmt_lin_turnover =
fit(inf_2cmt_lin_turnover, Subject.(sims), init_params(inf_2cmt_lin_turnover), FOCE())FittedPumasModel
Successful minimization: true
Likelihood approximation: FOCE
Likelihood Optimizer: BFGS
Dynamical system type: Nonlinear ODE
Solver(s): (Vern7,Rodas5P)
Log-likelihood value: -156.74137
Number of subjects: 10
Number of parameters: Fixed Optimized
0 15
Observation records: Active Missing
dv: 150 0
resp: 150 0
Total: 300 0
--------------------------
Estimate
--------------------------
tvcl 1.1682
tvvc 26.801
tvq 6.3267
tvvp 165.69
Ω_pk₁,₁ 0.17137
Ω_pk₂,₂ 0.091537
Ω_pk₃,₃ 0.055024
Ω_pk₄,₄ 2.9089e-74
σ_prop_pk 0.00084244
σ_add_pk 0.53685
tvturn 9.8483
tvebase 11.016
tvec50 0.28956
Ω_pd₁,₁ 0.025593
σ_add_pd 0.21128
--------------------------Calculating confidence intervals and Bootstrap
You can calculate confidence intervals using the infer function (by default 95% CIs):
infer_2cmt_lin_turnover = infer(fit_2cmt_lin_turnover)Asymptotic inference results
Successful minimization: true
Likelihood approximation: FOCE
Likelihood Optimizer: BFGS
Dynamical system type: Nonlinear ODE
Solver(s): (Vern7,Rodas5P)
Log-likelihood value: -156.74137
Number of subjects: 10
Number of parameters: Fixed Optimized
0 15
Observation records: Active Missing
dv: 150 0
resp: 150 0
Total: 300 0
------------------------------------------------
Estimate SE 95.0% C.I.
------------------------------------------------
tvcl 1.1682 NaN [NaN; NaN]
tvvc 26.801 NaN [NaN; NaN]
tvq 6.3267 NaN [NaN; NaN]
tvvp 165.69 NaN [NaN; NaN]
Ω_pk₁,₁ 0.17137 NaN [NaN; NaN]
Ω_pk₂,₂ 0.091537 NaN [NaN; NaN]
Ω_pk₃,₃ 0.055024 NaN [NaN; NaN]
Ω_pk₄,₄ 2.9089e-74 NaN [NaN; NaN]
σ_prop_pk 0.00084244 NaN [NaN; NaN]
σ_add_pk 0.53685 NaN [NaN; NaN]
tvturn 9.8483 NaN [NaN; NaN]
tvebase 11.016 NaN [NaN; NaN]
tvec50 0.28956 NaN [NaN; NaN]
Ω_pd₁,₁ 0.025593 NaN [NaN; NaN]
σ_add_pd 0.21128 NaN [NaN; NaN]
------------------------------------------------
Variance-covariance matrix could not be
evaluated. The random effects may be over-
parameterized. Check the coefficients for
variance estimates near zero.You can also convert the output to a DataFrame with the coeftable function:
coeftable(infer_2cmt_lin_turnover)| Row | parameter | estimate | se | relative_se | ci_lower | ci_upper |
|---|---|---|---|---|---|---|
| String | Float64 | Float64 | Float64 | Float64 | Float64 | |
| 1 | tvcl | 1.16817 | NaN | NaN | NaN | NaN |
| 2 | tvvc | 26.8008 | NaN | NaN | NaN | NaN |
| 3 | tvq | 6.32672 | NaN | NaN | NaN | NaN |
| 4 | tvvp | 165.695 | NaN | NaN | NaN | NaN |
| 5 | Ω_pk₁,₁ | 0.171366 | NaN | NaN | NaN | NaN |
| 6 | Ω_pk₂,₂ | 0.0915374 | NaN | NaN | NaN | NaN |
| 7 | Ω_pk₃,₃ | 0.0550237 | NaN | NaN | NaN | NaN |
| 8 | Ω_pk₄,₄ | 2.90889e-74 | NaN | NaN | NaN | NaN |
| 9 | σ_prop_pk | 0.000842441 | NaN | NaN | NaN | NaN |
| 10 | σ_add_pk | 0.536852 | NaN | NaN | NaN | NaN |
| 11 | tvturn | 9.84831 | NaN | NaN | NaN | NaN |
| 12 | tvebase | 11.0164 | NaN | NaN | NaN | NaN |
| 13 | tvec50 | 0.289555 | NaN | NaN | NaN | NaN |
| 14 | Ω_pd₁,₁ | 0.0255929 | NaN | NaN | NaN | NaN |
| 15 | σ_add_pd | 0.211277 | NaN | NaN | NaN | NaN |
The returned table also includes relative standard errors (RSEs). For inference with bootstrap-based confidence intervals you can use Bootstrap as an optional second positional argument in infer:
Pumas.Bootstrap — TypeBootstrap(; rng=Random.default_rng, samples=200, stratify_by=nothing, ensemblealg=EnsembleThreads())Constructs a Bootstrap object that allows the user to use bootstrapping with the supplies setting in infer function calls. See ?infer and https://docs.pumas.ai/stable/analysis/inference/ for more information about obtaining inference results.
The keyword arguments are as follows:
- rng: used to select a pseudo-random number generator to beused for the random resamples of the datasets.
- samples: controls the number of resampled
Populations. - resamples: the number of model parameter samples from the original sample distribution to re-sample (by weighed re-sampling without replacement). The number of resamples must be less than that of the original sample. An original sample-to-resample ratio of at least 5:1 is suggested.
stratify_bycontrol the stratification used in the re-sampling scheme. Should be a set to aSymbolwith the name of a covariate it is possible to stratify by a covariate with a finite number of possible values (e.g:studyfor a covariate namedstudy).ensemblealgcontrols the parallelization acrossfitcalls. Each fit is fitted using theensemblealgthat was used in the originalfit.
Performing the bootstrap for our model can be performed as:
infer_boot_2cmt_lin_turnover =
infer(fit_2cmt_lin_turnover, Bootstrap(; ensemblealg = EnsembleThreads()))Bootstrap inference results
Successful minimization: true
Likelihood approximation: FOCE
Likelihood Optimizer: BFGS
Dynamical system type: Nonlinear ODE
Solver(s): (Vern7,Rodas5P)
Log-likelihood value: -156.74137
Number of subjects: 10
Number of parameters: Fixed Optimized
0 15
Observation records: Active Missing
dv: 150 0
resp: 150 0
Total: 300 0
--------------------------------------------------------------------------------
Estimate SE 95.0% C.I.
--------------------------------------------------------------------------------
tvcl 1.1682 0.26444 [ 0.53796 ; 1.6056 ]
tvvc 26.801 6.5498 [ 19.663 ; 44.861 ]
tvq 6.3267 0.63479 [ 5.5201 ; 7.9791 ]
tvvp 165.69 27.436 [119.75 ; 226.47 ]
Ω_pk₁,₁ 0.17137 0.19149 [ 0.051904 ; 0.64125 ]
Ω_pk₂,₂ 0.091537 0.070162 [ 4.0211e-9 ; 0.22477 ]
Ω_pk₃,₃ 0.055024 0.033284 [ 5.8373e-9 ; 0.10965 ]
Ω_pk₄,₄ 2.9089e-74 3.9371e-90 [ 2.9089e-74; 2.9089e-74]
σ_prop_pk 0.00084244 4.2317e-7 [ 0.00084171; 0.00084318]
σ_add_pk 0.53685 0.025487 [ 0.48387 ; 0.58449 ]
tvturn 9.8483 0.51322 [ 8.4322 ; 10.511 ]
tvebase 11.016 0.5216 [ 10.117 ; 12.141 ]
tvec50 0.28956 0.030112 [ 0.24576 ; 0.3583 ]
Ω_pd₁,₁ 0.025593 0.0080927 [ 0.0067727 ; 0.039601 ]
σ_add_pd 0.21128 0.015438 [ 0.17598 ; 0.23344 ]
--------------------------------------------------------------------------------
Successful fits: 200 out of 200
Unique resampled populations: 200
No stratification.You can also convert the Bootstrap inference results to a DataFrame with coeftable:
coeftable(infer_boot_2cmt_lin_turnover)| Row | parameter | estimate | se | relative_se | ci_lower | ci_upper |
|---|---|---|---|---|---|---|
| String | Float64 | Float64 | Float64 | Float64 | Float64 | |
| 1 | tvcl | 1.16817 | 0.264438 | 0.22637 | 0.537957 | 1.60558 |
| 2 | tvvc | 26.8008 | 6.54983 | 0.24439 | 19.6627 | 44.8611 |
| 3 | tvq | 6.32672 | 0.634794 | 0.100335 | 5.52008 | 7.97907 |
| 4 | tvvp | 165.695 | 27.4359 | 0.165581 | 119.754 | 226.474 |
| 5 | Ω_pk₁,₁ | 0.171366 | 0.191492 | 1.11745 | 0.0519037 | 0.641253 |
| 6 | Ω_pk₂,₂ | 0.0915374 | 0.0701618 | 0.766483 | 4.02113e-9 | 0.224768 |
| 7 | Ω_pk₃,₃ | 0.0550237 | 0.0332838 | 0.6049 | 5.83728e-9 | 0.109651 |
| 8 | Ω_pk₄,₄ | 2.90889e-74 | 3.93713e-90 | 1.35348e-16 | 2.90889e-74 | 2.90889e-74 |
| 9 | σ_prop_pk | 0.000842441 | 4.23165e-7 | 0.000502308 | 0.000841715 | 0.000843182 |
| 10 | σ_add_pk | 0.536852 | 0.0254867 | 0.0474744 | 0.483865 | 0.584492 |
| 11 | tvturn | 9.84831 | 0.513219 | 0.0521124 | 8.43221 | 10.5107 |
| 12 | tvebase | 11.0164 | 0.521602 | 0.0473479 | 10.1171 | 12.1409 |
| 13 | tvec50 | 0.289555 | 0.0301123 | 0.103995 | 0.245757 | 0.358304 |
| 14 | Ω_pd₁,₁ | 0.0255929 | 0.00809269 | 0.316209 | 0.00677269 | 0.0396012 |
| 15 | σ_add_pd | 0.211277 | 0.0154376 | 0.073068 | 0.175978 | 0.233437 |
Conclusion
This tutorial covered the basic workflow of constructing a model, simulating observations from it, and conducting parameter estimation. You can also see subsequent tutorials that discuss the components in more detail, such as:
- More detailed treatment of specifying populations, dosage regimens, and covariates.
- Reading in dosage regimens and observations from standard input data.
- Fitting models with different estimation methods.